Automatic force field generation by self-learning hybrid Monte Carlo method (cooperation with NeuralMD)¶
With this function, a force field for NeuralMD is automatically generated from a single initial structure. In constant to the grand-project where the user generate training data, this function does not require the preparation of training data; only one initial structure is needed.
Note
To use this function, a separate NeuralMD license is required. Please refer to this description if the setup has not been completed yet.
Usage¶
1. Prepare initial structure¶
Open the project that will be used as the initial structure. Ensure the calculation engine is set to Q.E. and make any necessary adjustment to the SCF settings. These settings are applied to the first-principle calculations within the self-learning hybrid Monte Carlo method.
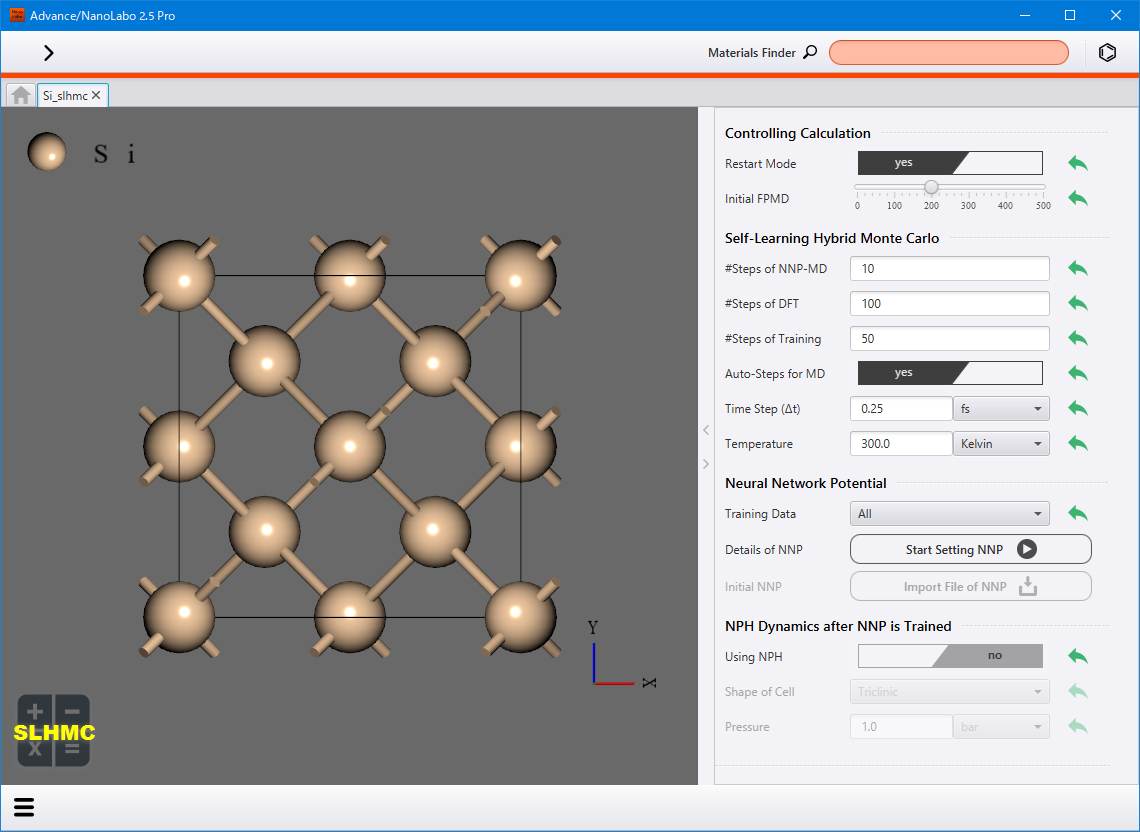
2. SLHMC setting¶
Click SLHMC from the lower-left menu ![]() . The settings screen for self-learning hybrid Monte Carlo method will be displayed. For detailed information about the setting items, please refer to the corresponding items in the configuration file description slhmc.prop in the NeuralMD document.
. The settings screen for self-learning hybrid Monte Carlo method will be displayed. For detailed information about the setting items, please refer to the corresponding items in the configuration file description slhmc.prop in the NeuralMD document.
When setting Method of Deformation of Cell to NPT Ensemble for Monte-Carlo or NPH-MD after NNP is Trained, cell deformation occurs during the SLHMC process.
Shape of Cell: Setting constraints for cell deformation
Isotropic
Allow isotropic deformation
Anisotropic
Allow stretching along x, y, z axes
Triclinic
Allow arbitrary deformation
Allow stretching along the 2 axes at the same ratio
Allow stretching along the 2 axes
Allow arbitrary deformations in a plane containing the 2 axes
Allow stretching along 1 axis
To use existing neural network force field as the initial force field instead of creating it from first-princible molecular dynamics calculations, set Initial FPMD to 0, and then click Import File of NNP at Initial NNP to select the ffield.sannp file.
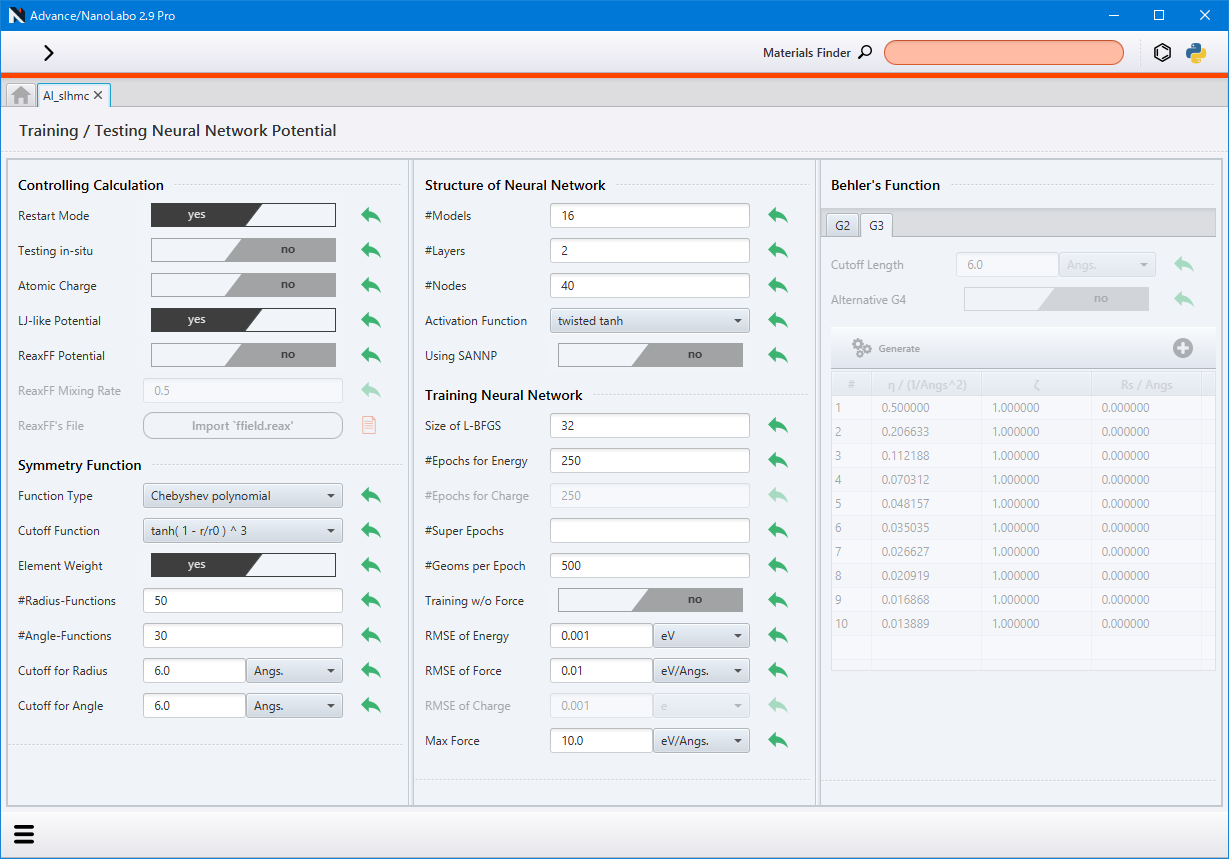
3. NNP setting¶
Click Details of NNP under Start Setting NNP to access the setting screen for the neural network potential. For detailed information about the setting items, please refer to the corresponding items in the configuration file description sannp.prop in the NeuralMD document.
If Training w/o Force is set to yes, the force component of the loss function is set to 0, and only energy is utilized for the training.
When the settings are complete, click Return to SLHMC from the lower-left menu ![]() to return to the original project screen.
to return to the original project screen.
4. Run¶
Click Run from the lower-left menu ![]() to initiate the process. In case of remote execution, the script editing screen for calling Quantum ESPRESSO, LAMMPS and NeuralMD will appear. Here, you can review and edit the content as needed. Afterward, click OK .
to initiate the process. In case of remote execution, the script editing screen for calling Quantum ESPRESSO, LAMMPS and NeuralMD will appear. Here, you can review and edit the content as needed. Afterward, click OK .
Note
Set #Processes to 1 for local calculation on Windows, as Windows version of NeuralMD does not currently support MPI parallelism. OpenMP parallelism is available.
Hint
You can achieve faster execution using GPU(s). The supported functions include training the neural network force field with Advance/NeuralMD (a license for the Advance/NeuralMD Pro edition is required) and performing molecular dynamics calculations using LAMMPS with a neural network force field.
(Linux only) To execute calculations locally, specify the number of GPU(s) to be used in the Number of GPU setting, found under . If multiple GPUs are utilized, MPI parallel processes are evenly distributed across the GPUs.
To execute calculations remotely, enable the GPU setting in the queue of the ssh server setting.
Note
The installation of a GPU driver is required prior to use. Since CUDA 11.4.4 is being utilized, a driver version of 470.82.01 or later is necessary.
If the system contains five or more elements, the weighted symmetry function must be used (set Element Weight to yes).
When you return to the tab, the result screen is displayed. You can check the status of first-principle molecular dynamics calculation (FPMD) used to create the initial neural network force field and the hybrid Monte Carle calculation (HMC). The neural network force field is updated in real-time, and you can determine the quality by evaluating if the Accepted Rate (that of Monte Carlo method) is closer to 1 and the mean absolute error (MAE) of energy is smaller. (However, since the number of steps in molecular dynamics calculations with neural network force fields may vary during the SLHMC process, these metrics may not always improve even if learning is successful.)
Click force-field to save trained neural network as a force field file for LAMMPS. Click train-data to save the generated training data.
Hint
To conduct molecular dynamics calculation using the created force field file, in a LAMMPS project, set the Type of Force Field to NeuralMD (or NeuralMD with charge) in the Force-Field setting screen, and select the created force field file in Potential File.
